Centre for Brain Development and Repair - CNS - Aditi Bhattacharya
Email: aditi at instem dot res dot in 
Aditi Bhattacharya
Decoding the proteome and translational control in Autism Spectrum Disorders and Intellectual Disability
Eukaryotic mRNA translation or protein synthesis is one of the most critical processes in an organism’s life, since proteins are the main executors of cellular function. Nowhere is the need for ‘on-demand’ protein synthesis felt more intensely than in neurons, where synaptic activity requires fast responses in a spatially-restricted manner. Protein synthesis is therefore considered an inescapable requisite to proper brain functioning. The proteome of any cell is “the set of expressed proteins at a given time, under defined conditions”. So it is easy to see that the proteome ‘fluctuates’ with different stimuli and measuring the proteome provides an unbiased snapshot of what proteins are deemed necessary to be expressed in a neuron/cell in response to said stimuli. Increasingly, a large number of psychiatric disorders are being reported to have deviations from ‘normal’ protein synthesis and so understanding what these changes are, will likely help to understand the disease phenotype better and devise novel biomarkers and therapeutic avenues.
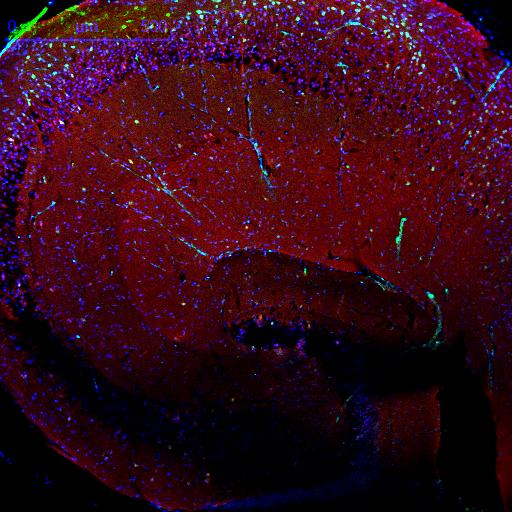 Image 1: Mouse hippocampal slice showing staining with FUNCAT (green), DAPI (blue) and tubulin (red).
Image 1: Mouse hippocampal slice showing staining with FUNCAT (green), DAPI (blue) and tubulin (red).
Genetic syndromes like Fragile X syndrome, Tuberous Sclerosis complex, Rett’s Syndrome etc. have been shown to have high co-incidence with autism spectrum disorders (ASD) and intellectual disability (ID). These conditions have also been demonstrated to have eccentric protein synthesis in neural cells. However the exact identity of proteins whose expression is dysregulated is still largely unknown. This is in part because tools to investigate proteomic changes in intact brain circuits is limited. I have co-developed an approach (BONLAC, Bowling-Bhattacharya et al., 2015) to address this problem by adapting existing cell biological tools like BONCAT/FUNCAT (Dieterich et al., 2006, 2010) and SILAC (Spellman et al.,2008) to look at proteomic changes in brain slices derived from rodent models of ASD/ID. My research interests are now to:
a) identify the circuits are differentially active when mice with ASD or ID, engage in specific ASD-like behaviors and if any proteins can be consistently identified to be expressed as a consequence of a certain behavior in these circuits
b) how the proteome changes when the brain develops in utero to early post-natal stages and how this is altered in ASD/ID
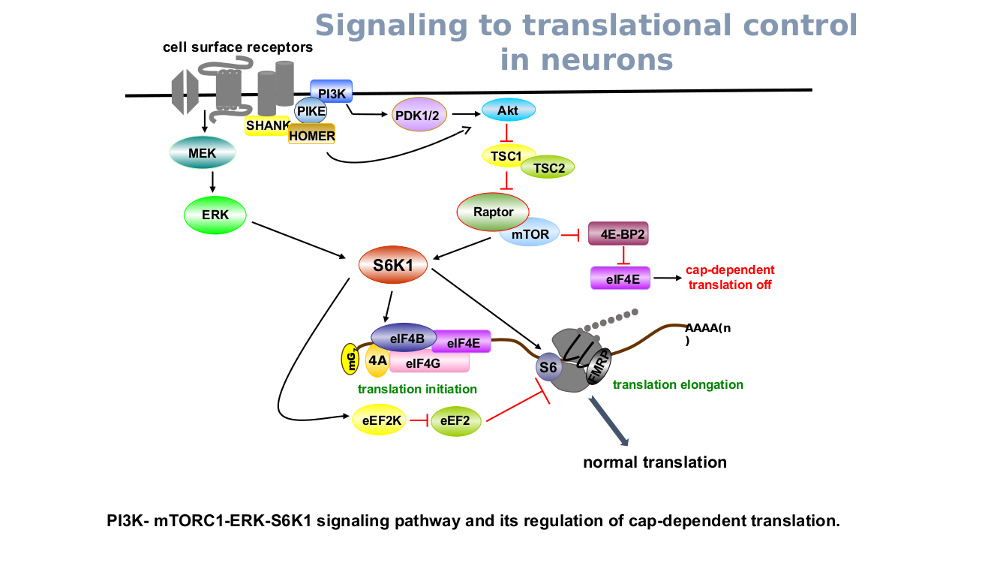
Another focus of my research will be to characterize the effect of de novo mutations in translation control molecules found in children with autism. As shown in Figure 2, this Akt-PI3K-activated pathway impinges on the mTORC1 , downstream of a myriad of cell surface signal transducers. Both mTORC1 and ERK signaling can activate S6K1 which promotes eukaryotic cap-dependent translation. The second arm of this signaling promotes eIF4E-eIF4G interactions via the inhibition of 4E-binding protein and stimulates translation initiation. Elevated levels of active mTORC1, ERK1/2, S6K1 and eIF4E lead to manifestation of
ASD-like phenotypes in mouse models (Hoeffer et al., 2008; Osterweil et al., 2010; Bhattacharya et al., 2012 and Santini et al., 2013). In addition recent whole exome studies (Iossifov et al., 2012 and 2015) have shown that children with ASD have aconcentration of mutations that are either spontaneous or inherited in the genomes as compared to their siblings or age matched controls. While the variants are now known at the genetic level, the impact that they have at the protein level is unknown. Beginning with mutations in S6K1 and mTOR, I intend to start unraveling the impact of these mutations on the structure-function of these kinases and what impact they have on basal and stimulus-triggered translation. their siblings or age matched controls. While the variants are now known at the genetic level, the impact that they have at the protein level is unknown. Beginning with mutations in S6K1 and mTOR, I intend to start unraveling the impact of these mutations on the structure-function of these kinases and what impact they have on basal and stimulus-triggered translation.
Selected Publications
Heather Bowling *, Aditi Bhattacharya* , Guoan Zhang , Joseph Z. Lebowitz , Danyal Alam , Rebecca Bisch , Peter T. Smith , Kent Kirschenbaum , Thomas A. Neubert, Christine Vogel , Moses V. Chao, and Eric Klann. (2015) Translational landscape of an adult rodent circuit in response to Brain Derived Neurotrophic Factor Stimulation. Neuropyschopharmacology, article in press, *equal first author. DOI: 10.1016/j.neuropharm.2015.07.017.
Daman Kumari*, Aditi Bhattacharya*, Jeff Nadel, Kirsten Moulton, Nicole Zeak, Anne Glicksman,Carl Dobkin, David Brick, Philip Schwartz, Carolyn B. Smith, Eric Klann, and Karen Usdin (2014) Identification of fragile X syndrome-specific molecular markers in human fibroblasts: a useful model to test the efficacy of therapeutic drugs. Human Mutation 35(12):1485-94, * equal first author. DOI:10.1002/humu.22699.
Heather Bowling, Guoan Zhang, Aditi Bhattacharya, Katrin Deinhardt, Charles A. Hoeffer, Thomas A. Neubert, Eric Klann, and Moses V. Chao. (2014) Antipsychotics enhance neuronal morphological complexity via mTORC1-dependent translation. Science Signaling epub-14 January 2014 Vol 7 Issue 308. DOI:10.1126/scisignal.2004331.
Aditi Bhattacharya, Hanoch Kaphzan, Amanda Alvarez-Dieppa, Jaclyn P. Murphy, Philippe Pierre, and Eric Klann. (2012) Correction of Molecular, Synaptic, Morphological, and Behavioral Phenotypes in Fragile X Syndrome Mice Via Genetic Removal of p70 S6 Kinase 1. Neuron, 76, 325-37. DOI: 10.1016/j.neuron.2012.07.022.
Aditi Bhattacharya and Eric Klann. (2012) The molecular basis of cognitive deficits in pervasive developmental disorders. Learn Mem.19, 434-43. 10.1101/lm.025007.111.
For full list: https://www.researchgate.net/profile/Aditi_Bhattacharya2