CNS, inStem – NYU Researchers Develop Method to Identify Newly Synthesized Proteins in Blood
We have all had blood tests in our lives that involved drawing a small quantity of blood from our veins and testing it for the levels of enzymes, vitamins, iron, and glucose. The blood draw remains the most commonly used biopsy for a multitude of measures for health and disease. However, what we test in these samples is usually levels of biomolecules that have been accumulating in our blood circulation over days to weeks. Accumulation can be by secretion of protein from cells in the liver, brain or kidney or metabolites being transported across organ systems. What we don’t normally end up measuring are the proteins that are dynamically made in blood cells even though these can be first responders in many immune challenge scenarios or can serve as excellent proxies to test if a systemic drug given for a particular disease is engaging the right signalling pathways. The stumbling block has been the lack of a method that enables proteins that at being synthesized at given point of time to be labelled and then detected accurately. Currently the approach is to isolate and culture specific blood cells and then carry out assays on them. If addressed, newly synthesized proteins in blood can serve as a new avenue of ‘biomarker’ discovery for a variety of conditions including slow progressing infectious disease, leukaemia, and potentially even neurological conditions.
An international collaboration between the laboratory of Prof. Eric Klann, New York University and Dr. Aditi Bhattacharya at the Center for Neurodevelopmental Synaptopathies (CNS), a joint programme between inStem, NCBS and the University of Edinburgh, has developed a simple solution to this problem. This study had been recently published in the Journal of Proteome Research. As the primary investigator of the project, Dr. Heather Bowling states “Measuring precise protein manufacturing within a specific time frame could reveal how blood maintains health and responds in disease conditions. The fact that this method can be used across multiple species from mice and rats to humans is especially beneficial for future research studies into blood response dynamics and to develop new therapeutics that target changes between health and disease.”
While the proof of concept studies were carried out at NYU, including a critical chemical biology validation at the laboratory of Prof. Kent Kirshenbaum, Dept of Chemistry, NYU; the confirmatory proteomic analysis was done at the mass spectrometry (MS) core at the Bangalore Life Science Cluster (BLiSC), of which inStem is an integral part. Dr. Chhaya Patole, a co-author in this study and in charge of the proteomic facility comments “MS core facility at BLiSC used the high end Thermo Fisher Scientific Orbitrap Fusion Tribrid that specializes in detecting the low abundant proteins from small input sample typically available for biopsies. In addition, we also optimized the data analysis work flow for high confidence detection of tagged newly synthesized proteins”.
Dr. Bhattacharya further quotes “the key component of this new technique required us to re-evaluate how a well-known chemical reaction called ‘click chemistry’ occurs in a sample enriched with ions and small molecule metabolites. Most of the traditional rules governing click chemistry had to be adjusted to achieve successful labelling of whole blood.”
The scientists hope that this technique will be adopted by groups across industry and academia to test in a variety of projects. While the most obvious first applications of this method would be in the field of developing new pharmacodynamic biomarkers in blood, authors can visualize its use in finding novel signatures for detection slow developing blood infections, companion biomarkers to cell cytometry based metrics for leukaemia or even standalone blood based biomarkers for rare neurological diseases.
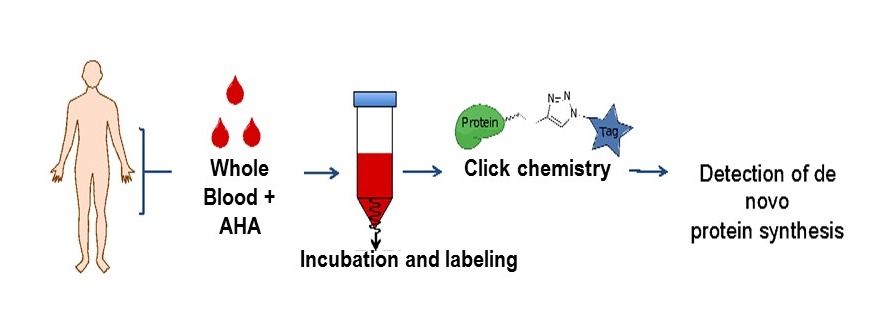
Schematic of the labelling technique to identify newly synthesized proteins in whole blood. Adapted from Bowling et al., 2020, J Proteome Res.
Reference:
Heather L. Bowling, Amanda Kasper, Chhaya Patole, Janani Priya Venkatasubramani, Sarah Parker Leventer, Erin Carmody, Kevin Sharp, Elizabeth Berry-Kravis, Kent Kirshenbaum, Eric Klann, and Aditi Bhattacharya, Aug 2020, Journal of Proteome Research
https://pubs.acs.org/doi/abs/10.1021/acs.jproteome.0c00299
Publication Date: August 4, 2020











